single cell RNAseq
Please read the following posts by Dave Tang. When I google, I always find his posts on top of the pages. Thanks for sharing your knowledge.
https://davetang.org/muse/2018/06/06/10x-single-cell-bam-files/ https://davetang.org/muse/2018/08/09/getting-started-with-cell-ranger/
From the 10x manual:
The final Single Cell 3’ Libraries contain the P5 and P7 primers used in Illumina bridge amplification PCR. The 10x Barcode and Read 1 (primer site for sequencing read 1) is added to the molecules during the GEMRT incubation. The P5 primer, Read 2 (primer site for sequencing read 2), Sample Index and P7 primer will be added during library construction
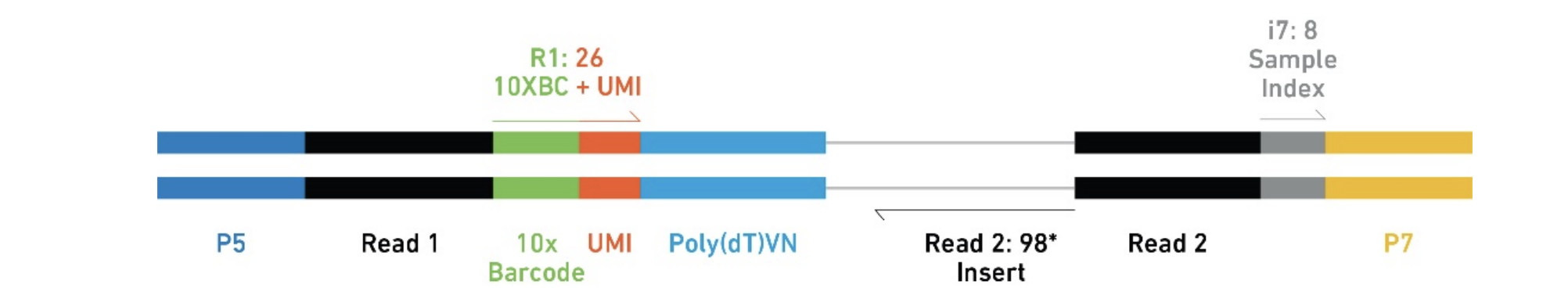
A Single Cell 3’ Library comprises standard Illumina paired-end constructs which begin and end with P5 and P7. The Single Cell 3’ v2 16 bp 10x Barcodes are encoded at the start of Read 1, while sample index sequences are incorporated as the i7 index read. Read 1 and Read 2 are standard Illumina sequencing primer sites used in paired-end sequencing. Read 1 is used to sequence the 16 bp 10x Barcode and 10 bp UMI, while Read 2 is used to sequence the cDNA fragment.
Each sample index provided in the Chromium i7 Sample Index Kit combines 4 different sequences in order to balance across all 4 nucleotides.
After cellranger mkfastq, three fastq.gz files will be produced: I1, R1 and R2. I1 is the 8 bp sample barcode, R1 is the 16bp feature barcode + 10 bp UMI, R2 is the reads mapped to the transcriptome.
Feature barcode whitelist can be found at the cellranger installation path: cellranger-2.1.0/cellranger-cs/2.1.0/lib/python/cellranger/barcodes/737K-august-2016.txt
single cell ATAC
after cellranger-atac mkfastq, there four fastq.gz files will be generated. I1, R1, R2 and R3.
I1 is the 8 bp sample barcode, R1 is the forward read, R2 is the 16 bp 10x feature barcode and R3 is the reverse read. Thanks Aditi Qamra for pointing it out.
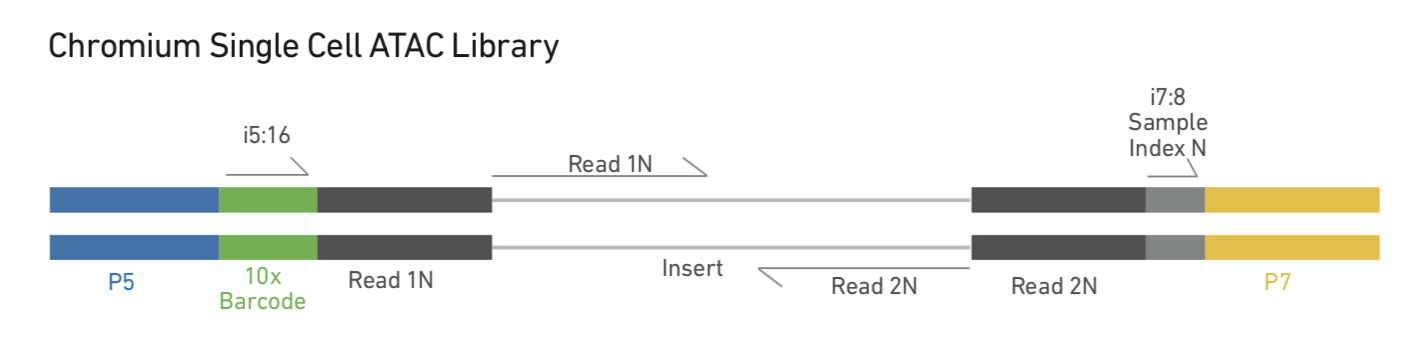
The 16bp feature barcode whitelist can be found at cellranger-atac installation path:cellranger-atac-1.0.1/cellranger-atac-cs/1.0.1/lib/python/barcodes/737K-cratac-v1.txt
Note that I have put the sample barcodes and feature barcodes files at https://osf.io/2z9gj/files/
Are the feature barcode whitelist the same for scRNAseq and scATAC?
In theory, 16bp barcode can have 4^16 (4,294,967,296) combinations, but you will want some diversities of the sequences to better distinguish 2 barcodes.Keep in mind that there are PCR or sequencing errors for the barcodes.
Both are 737K, but are the sequences the same?
cat 737K-august-2016.txt | sort | uniq > scRNAseq_barcode.txt
cat 737K-cratac-v1.txt | sort | uniq > scATAC_barcode.txt
wc -l scRNAseq_barcode.txt
737280 scRNAseq_barcode.txt
wc -l scATAC_barcode.txt
737280 scATAC_barcode.txt
## only 10812 cell barcodes are common
comm -12 scRNAseq_barcode.txt scATAC_barcode.txt | wc -l
10812
So, 10x uses quite different cell barcodes for scRNAseq and scATACseq applications.